AAPPred epitope prediction software based on the algorithm described in the article Prediction of linear B-cell epitopes using amino acid pair antigenicity scale.
The good statistical performance of the model was demonstrated in two benchmarks (Costa et al. 2013, Wang et al. 2018).
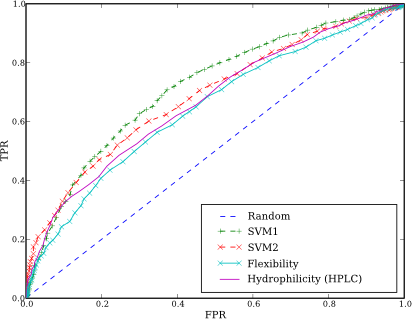
We have implemented two modes: first (SVM1) one is based on amino acids pair (AAP) frequencies and classical amino acids scales such as hydrophilicity, flexibility, etc. SVM2 uses only AAP frequencies to predict epitopes. The source code is available from the repository.
Our dataset contains not only publicly available data but also our proprietary experimental data.
This is ROC curve for our algorithm:

Please cite us: Ya.I. Davydov, A.G. Tonevitsky, 2009, published in Molekulyarnaya Biologiya, 2009, Vol. 43, No. 1, pp. 166–174.
 ), A. G. Tonevitsky
), A. G. Tonevitsky